Cardiac channelosomes: focus on the sodium channel Nav1.5
Numerous human diseases are caused by genetic or acquired ion channel dysfunction called channelopathies. Ion channels are membrane proteins allowing the passage of ions. The cardiac sodium ion channel Nav1.5, which gene is SCN5A, plays a pivotal role in such disorders. Nav1.5 channels are mainly expressed in the heart. Several hundred genetic variants in SCN5A were found in patients with a broad spectrum of cardiac manifestations, such as LQTS type 3, BrS, atrial fibrillation, and dilated cardiomyopathy. As with other proteins, the Nav1.5 channel interacts with many proteins, leading to its fine-tuning of expression, localization, and function. The work of our group is focused on deciphering which partner proteins interact with Nav1.5, leading to its proper function. The main technics used to perform such investigations are biochemical assays named co-immunoprecipitation (co-IP) experiments. However, the crucial step for this biochemical assay is the extraction of large proteins such as Nav1.5 α-subunit monomer (~220 KDa). In addition to their size, the embedment of those ion channels in a lipid bilayer, such as a plasma membrane, renders the extraction more difficult than cyto-soluble protein.
The homogenizer ‘Bioprep-24R’’ (ALLSHENG) that we were able to acquire, thanks to the UniBern Forschungsstiftungs’ grant, becomes essential for such investigation. The team members largely benefit from this new equipment to extract efficiently voltage-gated ion channels expressed in different organs and tissues. The homogenizer permits fast, effective, and reproducible homogenization, relating to the three-dimensional high-speed vibration and beating of grinding beads (glass beads, ceramic beads, steel balls, etc.).
We can now efficiently extract Nav1.5 channels from animal organs, a critical step to performing co-immunoprecipitation experiments.
PD Dr. Jean-Sébastien ROUGIER
Institute of Biochemistry and Molecular Medicine
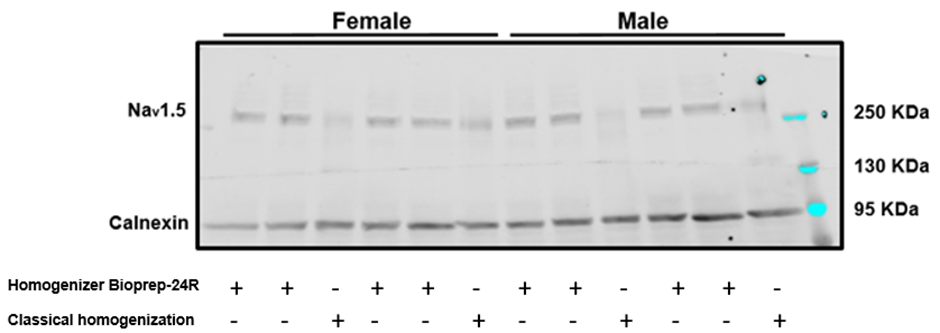
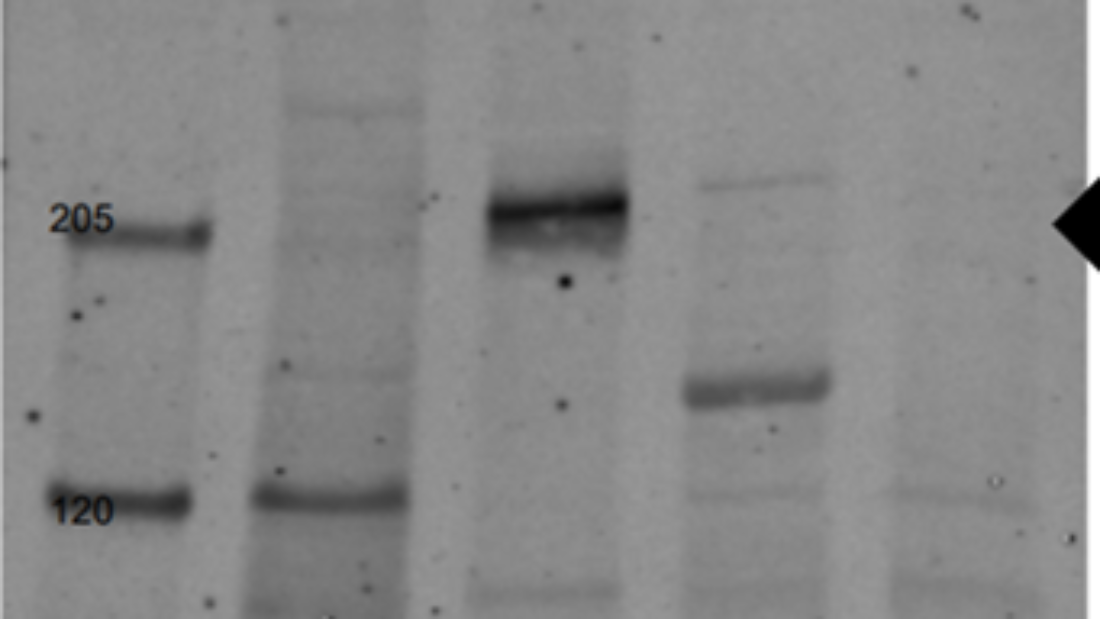
Western blot showing the efficiency of Nav1.5 extraction from murine hearts using either the classical homogenization procedure or the Bioprep-24R.
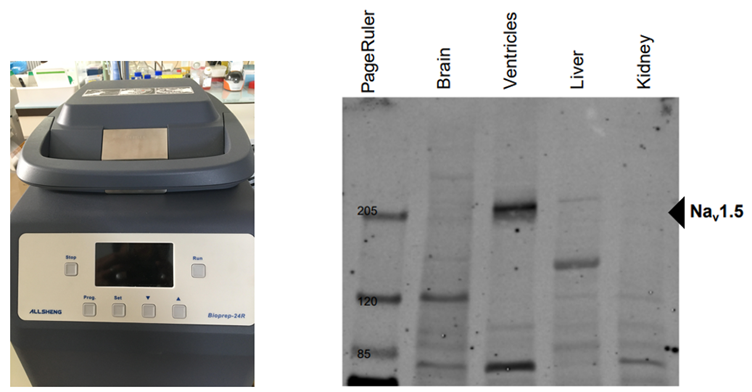
The homogenizer ‘Bioprep-24R’’ (ALLSHENG) and western blot showing the expression of Nav1.5 channel exclusively in the murine heart (ventricles).